Regarded as an aerobic, Gram-negative bacteria, Pseudomonas have been identified to display a wide variety of metabolic diversity, which enabled them to thrive in the ever-changing environment and occupy numerous ecological positions. Although they have been previously well known for causing severe infections in both animals and plants and thereby possessed negative reputations, they are, in fact, very effective in industries that utilize synthetic biology to produce complex chemicals that are rather challenging to synthesize using organic chemistry approaches. Also, as environmental pollution has emerged as a worldwide issue, it has become increasingly crucial to develop an effective way of purifying the sewage water. In this process, synthetic biology could be utilized to realize the bioremediation of the sewages that are polluted by organic matters. Pseudomonas is known to be an important constituent of biofilms, which is composed of multiple types of microbes and numerous organic matters. A reasonable amount of biofilm could be used to break down organic matters in sewages effectively and precisely into nonhazardous molecules. However, when they overgrown in the pipelines of water transporting systems, they could possibly block the water transporting system. Thus, it is crucial to control the amounts of biofilms within a reasonable amount. Phages targeting multiple microorganisms could be utilized to degrade biofilms. However, since the multiplication of phage can hardly be estimated, the abuse of phage infection could possibly cause severe damage to the preexisting biofilm systems. Thus, in order to realize precise and dynamic control over the infection of phage and the population size of the biofilm, a small molecule drug is required to help the bacterium to resist the infection of the bacterium.
Proof of Concept
Raising the scientific question: all about Pseudomonas & phage infection
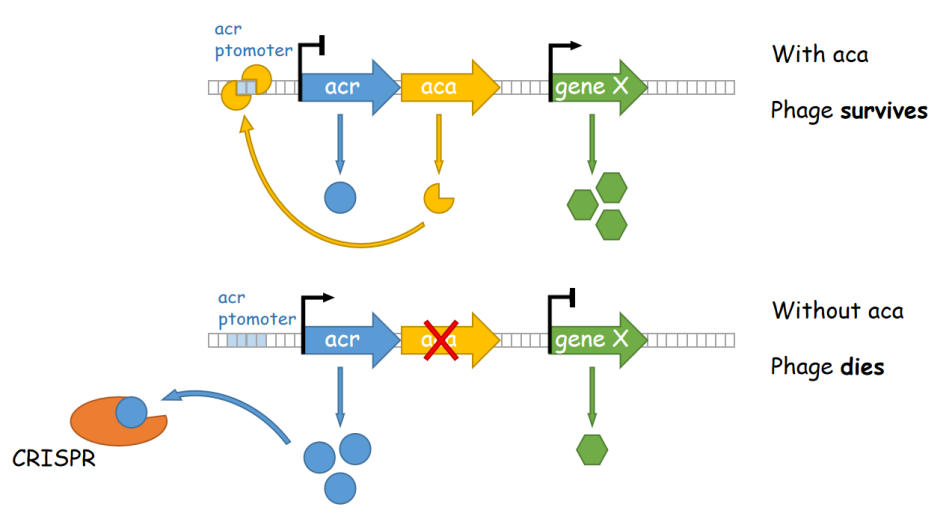
Although the utilization of Pseudomonas in synthetic biology, as well as biological degradation, could be rather effective and satisfying, certain problems still existed. Both these industrial applications required a mass amount of bacterial cultures, which in fact suffer severely from phage infection. Take E. coli and A. tumefaciens, which are two important tools utilized in laboratories to express proteins and to create transgenic plant seedlings as an example, the mass culture of these bacteria often encounter the infection of phages, which spreads quickly and would result in a clear, transparent culture media with no bacterium. This phenomenon also appears in industrial culturing of Pseudomonas, and there has not yet been any reported solution to this issue. According to current researches, phage infection of Pseudomonas is majorly mediated via an anti-CRISPR mechanism utilized by phages to conquer the line of defense of the bacterium. In this process, phages express anti-CRISPR (ACR) proteins to inhibit the CRISPR-Cas system, preventing their own genomes from being destroyed. Most acr genes are adjacent to anti-CRISPR-associated (aca) genes, which encode proteins that bind to DNA and inhibit downstream transcriptional activities. Thus, after phage DNA injection, Aca proteins within the bacterium would bind to the promoter regions of highly transcribed acr genes and thereby inhibit their transcription, which prevents the disruption of downstream gene transcription mediated by Acr proteins, thereby promoting the phage’s spreading and damaging the cells.
Figure 1. A working model of aca-medaited phage resistance
Combining the aforementioned discoveries and researches, here we propose a combination of structural biology and chemical genetics screening to provide a solution for phage-mediated cell damage in synthetic biology applications. Moreover, via this approach, we intend to provide a practical strategy to develop small molecule inhibitors of transcription factors and thereby enable us to artificially realize precise spatiotemporal control over biological systems utilized in synthetic biology.
Providing a possible solution: Combining structural biology & Target directed chemical screening
In this study, we initially purified high-quality Aca1 proteins (in dimer form) and co-crystallized them with their recognizing DNA fragments. Precise crystal structures were obtained by analyzing the X-ray diffraction pattern of the complex. With this structure, an in silico docking utilizing the well-characterized molecular docking software Molecular Operating Environment (MOE) over 5,800 commercially available small molecule compounds were performed. In this docking process, we set up two potential sites of interaction, one site lied between the hydrophobic surface of Aca1 protein, aiming to select chemical candidates that could adhere to the surface and therefore work as protein-protein interaction (PPI) inhibitors. The second location was set at the protein-DNA interacting site, aiming to prevent the interaction between Aca1 protein and its DNA binding site (PDI-inhibitors), and thereby functionally disable these proteins.
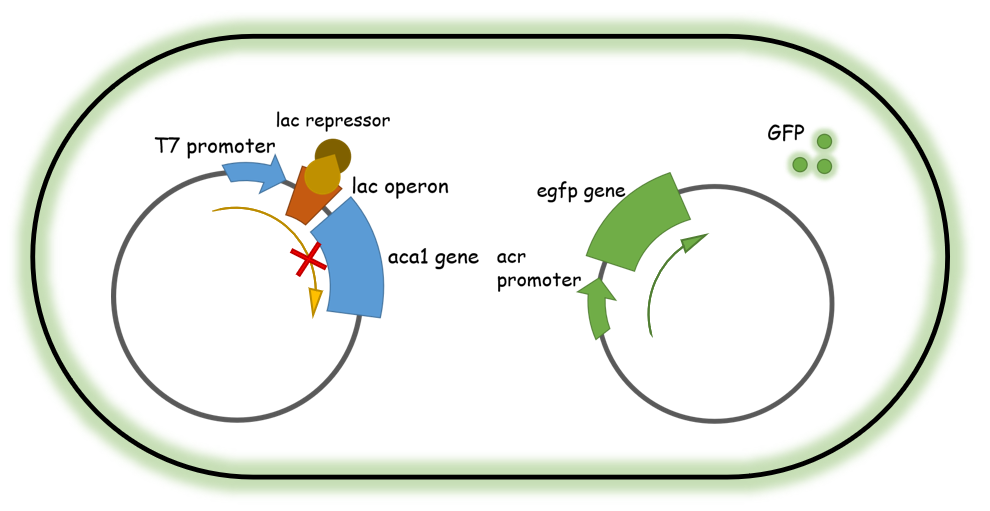
After several potential candidates were obtained, we then constructed a GFP-based reporter system (genetically modified E. coli strains) that contained two plasmids: one containing a T7 promoter-driven Aca1 protein, controlled via a lac operon, that generates functional Aca1 proteins upon IPTG treatment. Another plasmid is an eGFP protein fused after an Aca1-binding promoter sequence that is constitutively expressed in the absence of Aca1 and could be robustly inhibited when abundant Aca1 is present. Utilizing this system, the expression of the eGFP protein is suppressed in the cell since a large amount of Aca1 transcriptional inhibitor is present. Therefore, if a functional Aca1-inhibiting small molecule compound is added into the system, it would release the Aca1 proteins from the eGFP promoter and therefore generate detectable green fluorescence signals in the cell culture. This system could be utilized in high-throughput screening over effective chemicals since they are easy to detect/evaluate and convenient to perform.
Figure 2. Illustration of the two-plasmid system
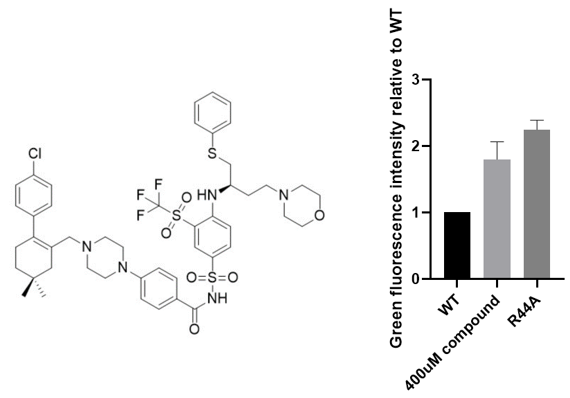
Later, we tested the top 20 candidates (each for PDI-inhibitors and PPI-inhibitors) with the highest potential binding affinity and obtained one chemical, Navitoclax, that displayed biological function within the reporter system. As a positive control, a mutated version (loss-of-function) of Aca1, whose Arginine residue at position 44 was altered to an Alanine residue, was introduced into the system. It could be observed that the addition of the compound induced a significant increase in the fluorescence intensity, even comparable to the mutation of the protein.
Figure 3. The chemical structure and inhibitory effect of Navitoclax
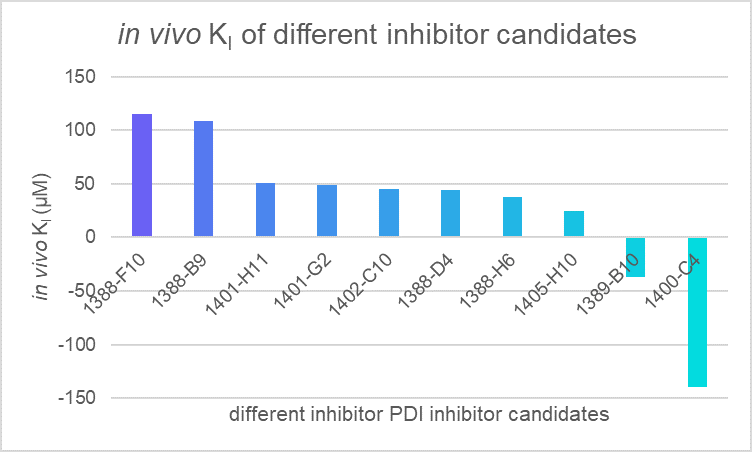
Also, we developed an evaluation system that could transform the fluorescence intensity signal into the content of inhibition. Using a constant named in vivo K1, we evaluate the in vivo efficiency of the potential inhibitors for protein-DNA interaction (PDI) as follows:
(1) ![]()
where KI is in vivo inhibitory constant of a specific PDI inhibitor, c(PDI-inhibitor) is the concentration of the PDI inhibitor in the system, I1 is the strength of the fluorescence in the positive control, I0 is the strength of the fluorescence in negative control and I is the strength of the fluorescence in the experimental group.
Figure 4. The estimated in vivo K1 of multiple chemical candidates
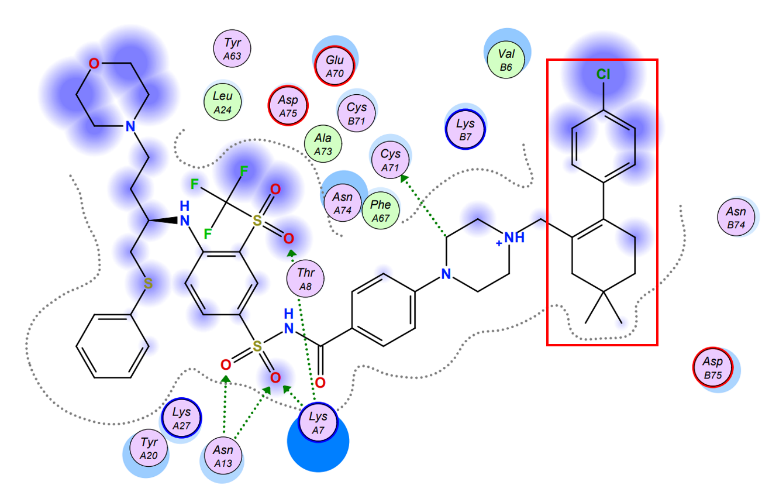
To maximize the efficiency of the chemical compound, we brought up several possible strategies, initially, the chemical is poorly soluble in the water, thus chemical modifications could be made to increase its hydrophilicity by removing certain hydrophobic residues. In order to make precise modifications, we estimated the potential interactions between Navitoclax and Aca1 amino acid residues and discovered that the aromatic tail of navitoclax displayed no potential interactions with the amino acid residues, and was responsible for the poor solubility of the compound. Therefore, utilizing synthetic chemistry, we designed a few more chemicals that structurally mimicked navitoclax but possessed higher solubility and binding affinity, which enabled them to be better candidates compared to navitoclax.
Figure 5. The estimated model of interaction between navitoclax & Aca1
Thus, combining structural biology and chemical genetics screening, we developed a novel small molecule inhibitor, navitoclax, that could inhibit the transcriptional function of Aca1, thereby assist Pseudomonas to defend against phage infection. In this way, when the biofilm overgrows in the sewage transporting pipeline, we will be able to apply phages to infect and remove the excess biofilm. As the superfluous bacterium is eliminated by phages, we could apply the small molecule drug to protect the bacterium from being excessively damaged by the application of phages. In all, since we are able to control the application time window and the concentration of the applied molecule, we are able to exert dynamic control over the growing status of the biofilms growing in the pipeline and thereby assist to deal with the city sewages using biological control.